
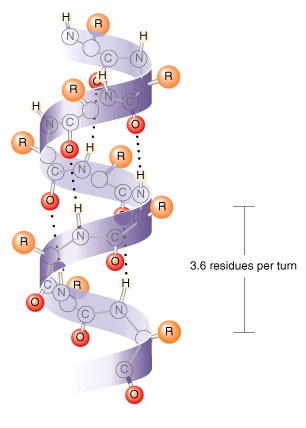
: 39 Most importantly, the N-H group of an amino acid forms a hydrogen bond with the C=O group of the amino acid three residues earlier this repeated i + 3 → i hydrogen bonding defines a 3 10-helix. Each amino acid corresponds to a 120° turn in the helix (i.e., the helix has three residues per turn), and a translation of 2.0 Å (0.20 nm) along the helical axis, and has 10 atoms in the ring formed by making the hydrogen bond. The amino acids in a 3 10-helix are arranged in a right-handed helical structure. Longer sections, in the range of seven to eleven residues, have been observed in the voltage sensor segment of voltage-gated potassium channels in the transmembrane domain of certain helical proteins. They are almost always short sections, with nearly 96% containing four or fewer amino acid residues, : 44 appearing in places such as the "corners" where α-helices change direction in the myoglobin structure, for example. The 3 10 helix is now known to be the third principal structure to occur in globular proteins, after the α-helix and β-sheet. The 3 10 helix was eventually confirmed by Kendrew in his 1958 structure of myoglobin, and was also found in Perutz' 1960 determination of the structure of haemoglobin and in subsequent work on both its deoxygenated and oxygenated forms.
ALPHA HELIX FREE
Those structures in which all available NH and CO groups are hydrogen bonded are inherently more probable, because their free energy is presumably lower.These form helical arrangements that cannot be uncoiled without breaking the hydrogen bonds. The chains are held together by hydrogen bonding between the hydrogen and oxygen atoms of different by nearby amide (peptide) links formed as the amino acids condense to form the polypeptide chain.The principles applied in the 1950 paper to theoretical polypeptide structures, true of the 3 10 helix, included: Perutz' confirmation of the alpha helix structure was published in Nature shortly afterwards. Within a few hours, he had the evidence to confirm the alpha helix, which he showed to Bragg first thing on Monday. Later that day, an idea for an experiment to confirm Pauling's model occurred to Perutz, and he rushed to the lab to carry it out. In contrast to Kendrew's and my helices, theirs was free of strain all of the amide groups were planar and every carbonyl group formed a perfect hydrogen bond with an amino group four residues further along the chain. I was thunderstruck by Pauling and Corey's paper. Perutz describes in his book I wish I'd made you angry sooner the experience of reading Pauling's paper one Saturday morning: Pauling was highly critical of the helical structures proposed by Bragg, Kendrew, and Perutz, taking a triumphal tone in declaring them all implausible. Watson's publication of the DNA double helix. The following year, Linus Pauling predicted both of those motifs, the alpha helix and the beta sheet, in work which is now compared in significance to Francis Crick and James D.
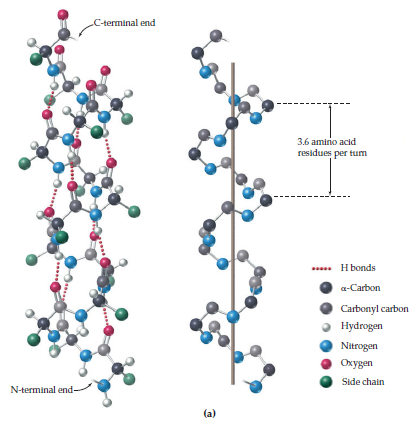
Their proposals included what is now known as the 3 10 helix, but did not include the two most common structural motifs now known to occur. Together with Lawrence Bragg and John Kendrew, Perutz published an exploration of polypeptide chain configurations in 1950, based on cues from noncrystalline diffraction data as well as from small molecule crystal structures such as crystalline found in hair. Max Perutz, the head of the Medical Research Council Laboratory of Molecular Biology at the University of Cambridge, wrote the first paper documenting the elusive 3 10-helix. Because of the α-helices tendency to consistently fold and unfold, it has been proposed that the 3 10-helix serves as an intermediary conformation of sorts, and provides insight into the initiation of α-helix folding. 3 10-helices constitute nearly 10–15% of all helices in protein secondary structures, and are typically observed as extensions of α-helices found at either their N- or C- termini. Of the numerous protein secondary structures present, the 3 10-helix is the fourth most common type observed following α-helices, β-sheets and reverse turns. Note that the sidechains point slightly downwards, i.e., towards the N-terminus.Ī 3 10 helix is a type of secondary structure found in proteins and polypeptides. The protein chain runs upwards, i.e., its N-terminus is at the bottom and its C-terminus at the top of the figure. Two hydrogen bonds to the same peptide group are highlighted in magenta the oxygen-hydrogen distance is 1.83 Å (183 pm). Side view of a 3 10-helix of alanine residues in atomic detail.


 0 kommentar(er)
0 kommentar(er)
